Summary
Background
Triple-negative breast cancer (TNBC), recognised as the most aggressive subtype of breast cancer, has a high recurrence rate and poor treatment outcomes. Current diagnosis relies heavily on immunohistochemistry, which can be time-consuming and costly, while prognostic stratification remains limited by traditional clinicopathological features. This study aimed to develop and validate an artificial intelligence (AI)-powered TNBC identification and prognosis prediction (TRIP) system using haematoxylin and eosin (H&E)-stained pathology images.
Methods
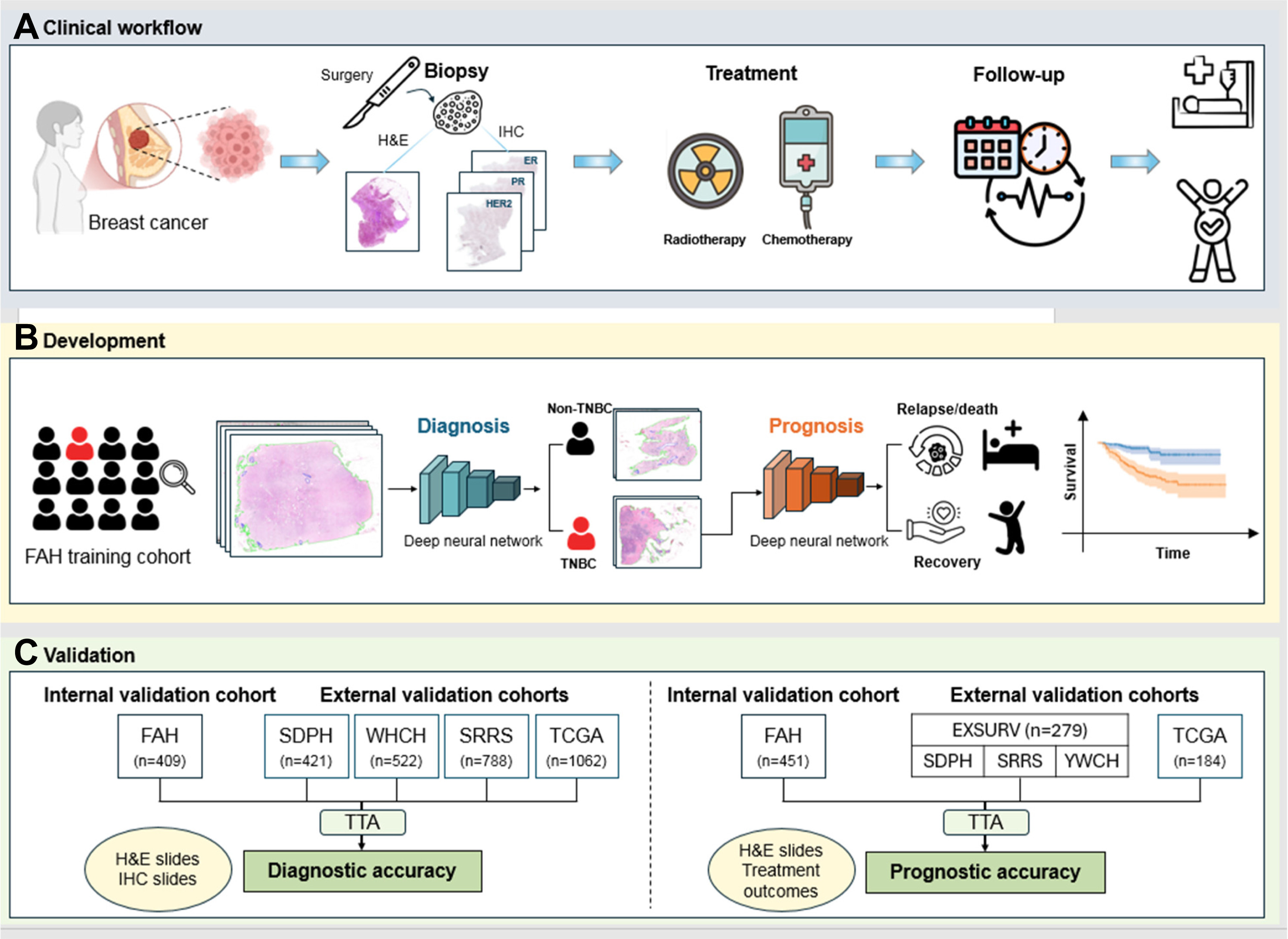
In this multicentre retrospective study, we analysed haematoxylin and eosin (H&E)-stained whole slide images (WSIs) of 2045 patients with breast cancer from The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China (FAH) between June 1, 2007 and December 31, 2022. Among these, 451 patients with TNBC had follow-up outcomes on disease-free survival and overall survival. Patients were excluded if they had other synchronous malignant neoplasms within five years or had previously received neoadjuvant chemotherapy. We developed and validated a deep learning system, TRIP, to classify TNBC versus other breast cancer subtypes and predict the cancer disease-free survival and overall survival on the FAH cohort. We employed the area under the receiver operating characteristic curve (AUC) to evaluate TNBC identification performance, and the concordance index (C-index) to evaluate the performance of disease-free survival and overall survival predictions. Beyond internal validation, the system was externally evaluated on independent retrospective cohorts from four tertiary hospitals in Eastern and Central China (Shandong Provincial Hospital [SDPH], Sir Run Run Shaw Hospital of Zhejiang University [SRRS], Yiwu Central Hospital [YWCH], and The Central Hospital of Wuhan [WHCH]) between June 26, 2013 and December 31, 2024, and The Cancer Genome Atlas (TCGA) dataset between January 1, 1988 and December 31, 2013, comprising 2793 cases for TNBC identification and 463 cases for prognosis prediction. Model interpretability was enhanced using pathology heatmaps, and multi-omics analysis was conducted to explore TNBC heterogeneity.
Findings
The deep-learning model achieved an AUC of 0.980, 95% Confidence Interval (CI): 0.958–0.996, for identifying TNBC in the internal cohort, and AUCs of 0.916 (95% CI: 0.848–0.959), 0.936 (95% CI: 0.907–0.962), 0.860 (95% CI: 0.779–0.930), and 0.890 (95% CI: 0.841–0.929) in the SDPH, SRRS, WHCH, and TCGA external validation cohorts, respectively. Moreover, it effectively predicted disease-free survival with a C-index of 0.747 ± 0.070 (95% CI: 0.617–0.852) in the internal cohort and C-indices of 0.731 ± 0.047 (95% CI: 0.623–0.839) and 0.732 ± 0.043 (95% CI: 0.621–0.840) in the SDPH, SRRS, YWCH cohorts combined (EXSURV), and the TCGA cohort, respectively, and overall survival with a C-index of 0.744 ± 0.075 (95% CI: 0.602–0.865) in the internal cohort and C-indices of 0.720 ± 0.034 (95% CI: 0.566–0.865) and 0.721 ± 0.030 (95% CI: 0.625–0.818) in the EXSURV and TCGA cohorts, respectively. Heatmaps revealed key histologic features of TNBC, including nuclear atypia, necrosis, and immune-cold microenvironments in aggressive cases, while lymphoplasmacytic infiltration indicated better prognosis. Multi-omics analysis identified three molecular subtypes with distinct immune and pro-tumour signalling profiles, supporting the TRIP system’s prognostic accuracy in alignment with molecular evidence.
Interpretation
TRIP is an effective and interpretable artificial intelligence system that demonstrates strong performance in identifying TNBC and predicting its disease-free survival and overall survival. However, the current findings are primarily derived from post-surgical tissue data and do not incorporate clinical variables, which limits the system’s immediate applicability in pre-operative settings. To overcome these constraints, future prospective studies should be conducted to validate its clinical utility, particularly in pre-operative and broader real-world scenarios.
Downloads
Full paper: click here
PyTorch code: click here
Reference
@article{zhang2025development,
title={Development and validation of an artificial intelligence system for triple-negative breast cancer identification and prognosis prediction: a multicentre retrospective study},
author={Zhang, Xiu-Ming and Zhou, Hua-Jun and Chen, Qing and Wang, Xi and Fu, Yu-Juan and Jin, Cheng and Zhou, Feng-Tao and Wang, Jing-Ping and Cai, Qiu-Yu and Wang, Ji-Li and others},
journal={eClinicalMedicine},
volume={89},
pages={103557},
year={2025},
publisher={Elsevier}
}