Abstract
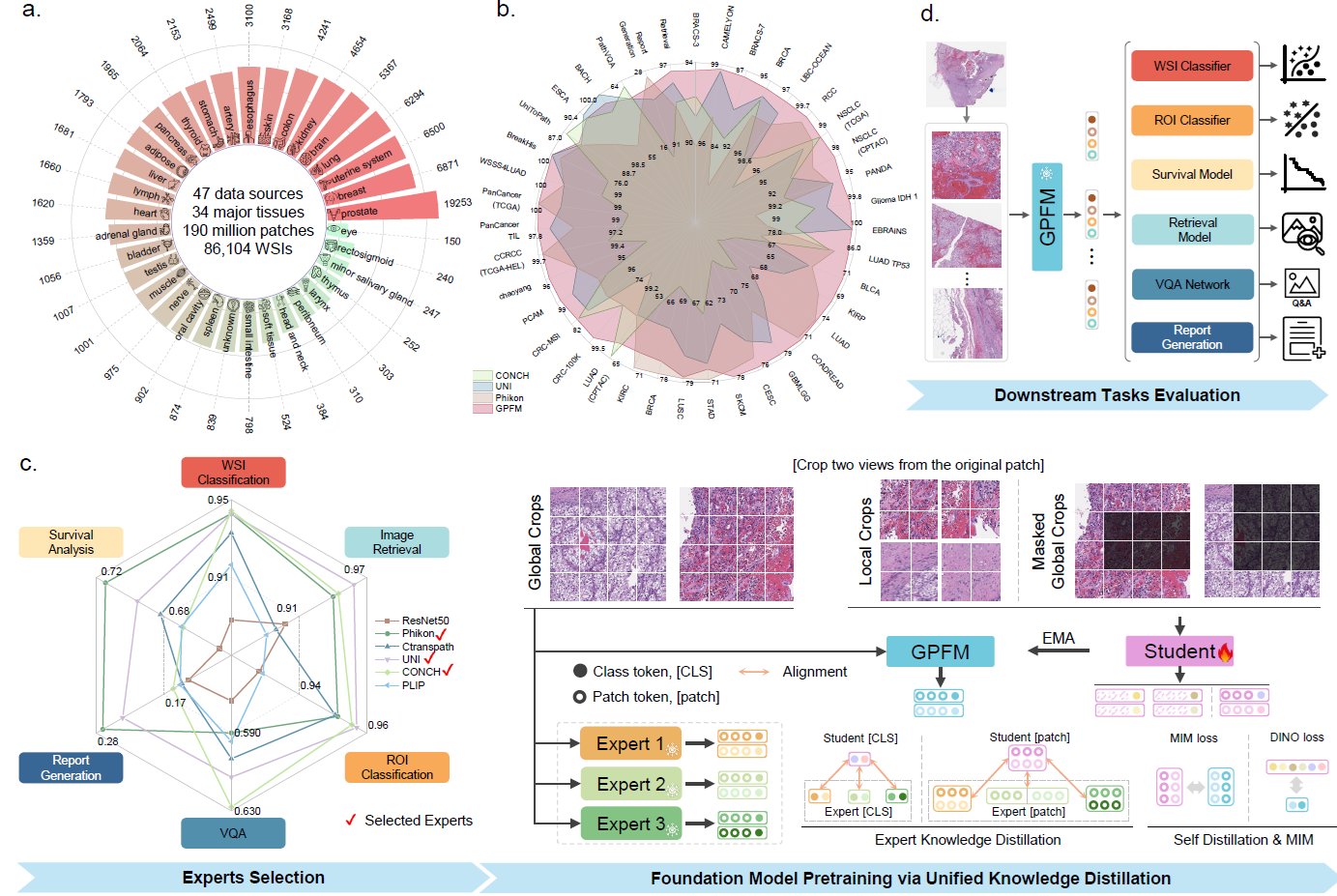
Foundation models pretrained on large-scale datasets are revolutionizing the field of computational pathology (CPath). The generalization ability of foundation models is crucial for the success in various downstream clinical tasks. However, current foundation models have only been evaluated on a limited type and number of tasks, leaving their generalization ability and overall performance unclear. To address this gap, we established a most comprehensive benchmark to evaluate the performance of off-the-shelf foundation models across six distinct clinical task types, encompassing a total of 72 specific tasks, including slide-level classification, survival prediction, ROI-tissue classification, ROI retrieval, visual question answering, and report generation. Our findings reveal that existing foundation models excel at certain task types but struggle to effectively handle the full breadth of clinical tasks. To improve the generalization of pathology foundation models, we propose a unified knowledge distillation framework consisting of both expert and self-knowledge distillation, where the former allows the model to learn from the knowledge of multiple expert models, while the latter leverages self-distillation to enable image representation learning via local-global alignment. Based on this framework, we curated a dataset of 96,000 whole slide images (WSIs) and developed a Generalizable Pathology Foundation Model (GPFM). This advanced model was trained on a substantial dataset comprising 190 million images extracted from approximately 72,000 publicly available slides, encompassing 34 major tissue types. Evaluated on the established benchmark, GPFM achieves an impressive average rank of 1.6, with 42 tasks ranked 1st, while the second-best model, UNI, attains an average rank of 3.7, with only 6 tasks ranked 1st. The superior generalization of GPFM demonstrates its exceptional modeling capabilities across a wide range of clinical tasks, positioning it as a new cornerstone for feature representation in CPath.
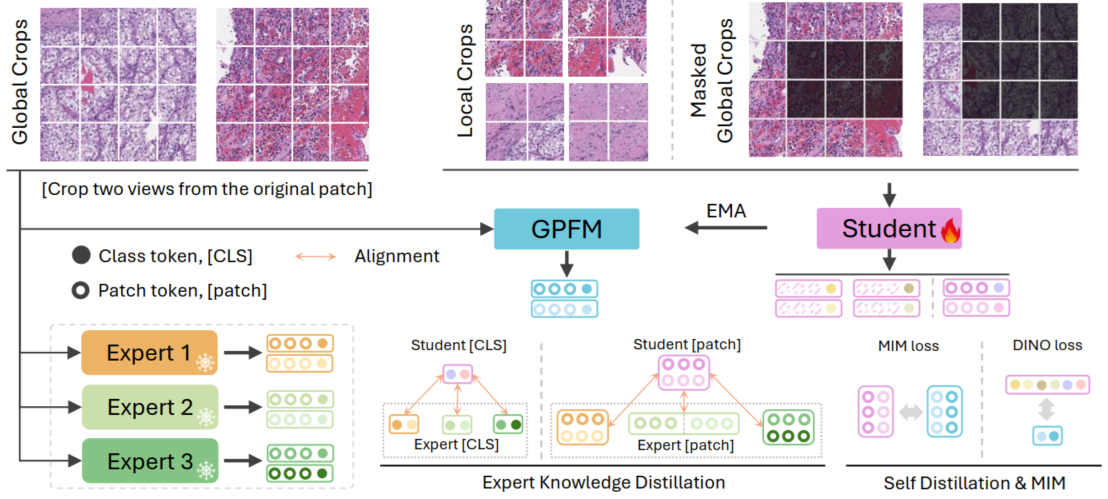
Schematic Diagram of the Proposed Method
We firstly choose the SoTA models from the off-the-shelf expert models, and then perform self-distillation to learn the local-global alignment. Finally, we obtain the unified knowledge distillation framework and train the GPFM on over 190 million pathology images.
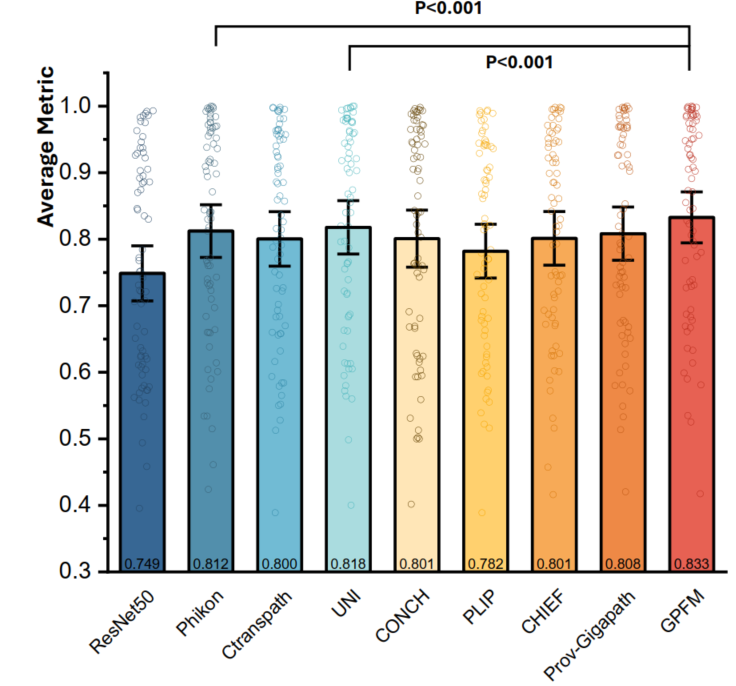
Ultimately, we evaluate the GPFM on the established benchmark containing 72 tasks and achieve state-of-the-art performance.
For the technical details, please refer to the original paper.
Downloads
Full Paper: click here
Supplementary: click here
Peer Review History: click here
PyTorch Code: click here
Reference
@article{ma2025generalizable,
title={A generalizable pathology foundation model using a unified knowledge distillation pretraining framework},
author={Ma, Jiabo and Guo, Zhengrui and Zhou, Fengtao and Wang, Yihui and Xu, Yingxue and Li, Jinbang and Yan, Fang and Cai, Yu and Zhu, Zhengjie and Jin, Cheng and others},
journal={Nature Biomedical Engineering},
pages={1--20},
year={2025},
publisher={Nature Publishing Group UK London}
}