Abstract
Fine-grained classification of whole slide images (WSIs) is essential in precision oncology, enabling precise cancer diagnosis and personalized treatment strategies. The core of this task involves distinguishing subtle morphological variations within the same broad category of gigapixel-resolution images, which presents a significant challenge. While the multi-instance learning (MIL) paradigm alleviates the computational burden of WSIs, existing MIL methods often overlook hierarchical label correlations, treating fine-grained classification as a conventional multi-class task. Introducing hierarchical information can enhance classification performance by leveraging the inherent relationships between different levels of labels, thus providing a more structured and informative learning process. To overcome these limitations, we introduce a novel hierarchical multi-instance learning (HMIL) framework. HMIL incorporates a class-wise attention mechanism that aligns hierarchical information at both the instance and bag levels. Furthermore, we introduce supervised contrastive learning to enhance the discriminative capability for fine-grained classification and a curriculum-based dynamic weighting module to adaptively balance the hierarchical feature during training. Extensive experiments on our large-scale cytology cervical cancer (CCC) dataset and two public histology datasets, BRACS and PANDA, demonstrate the state-of-the-art performance of our HMIL framework. Our source code is available at https://github.com/ChengJin-git/HMIL.
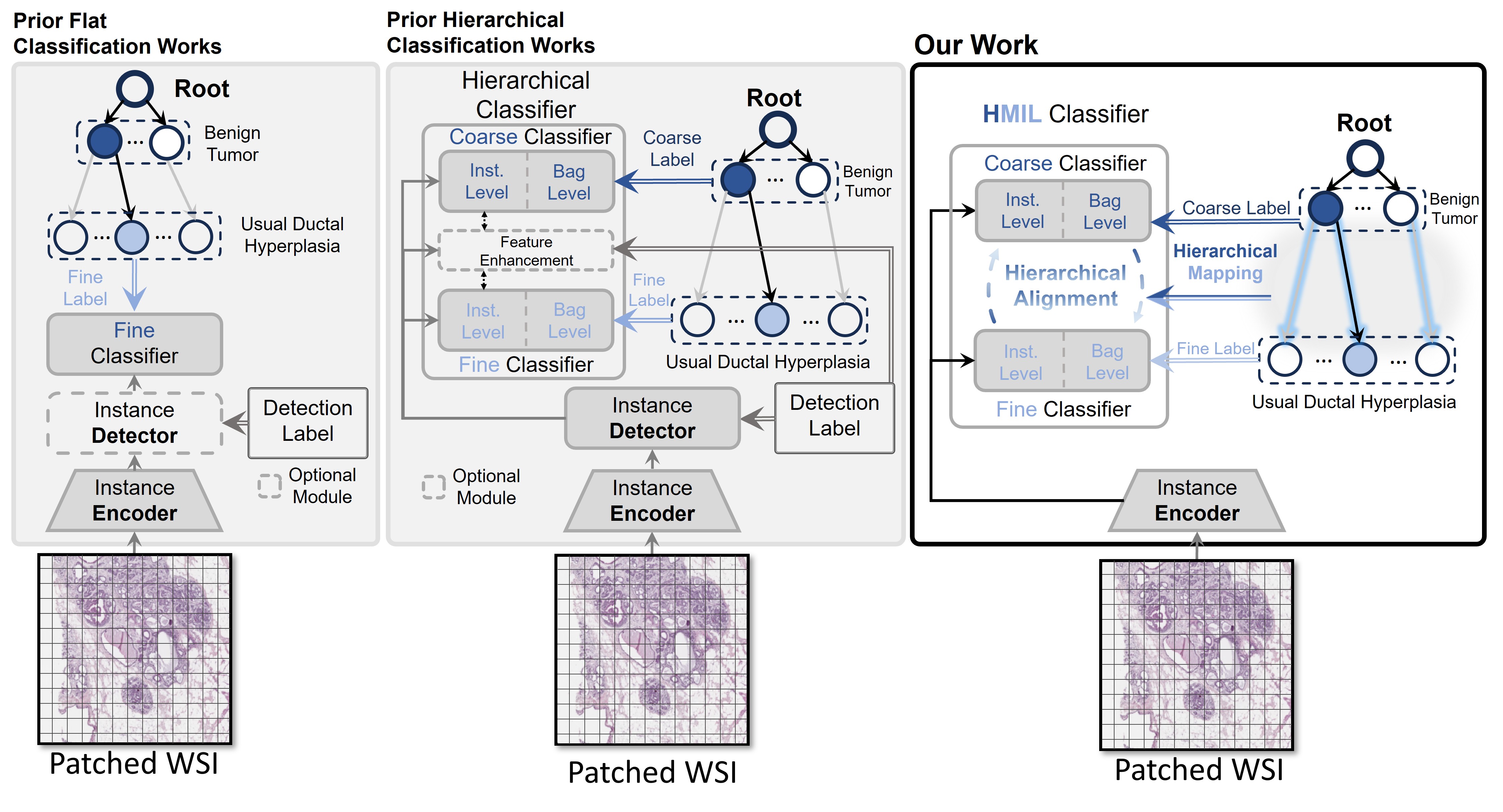
Schematic Diagram of the Proposed Method
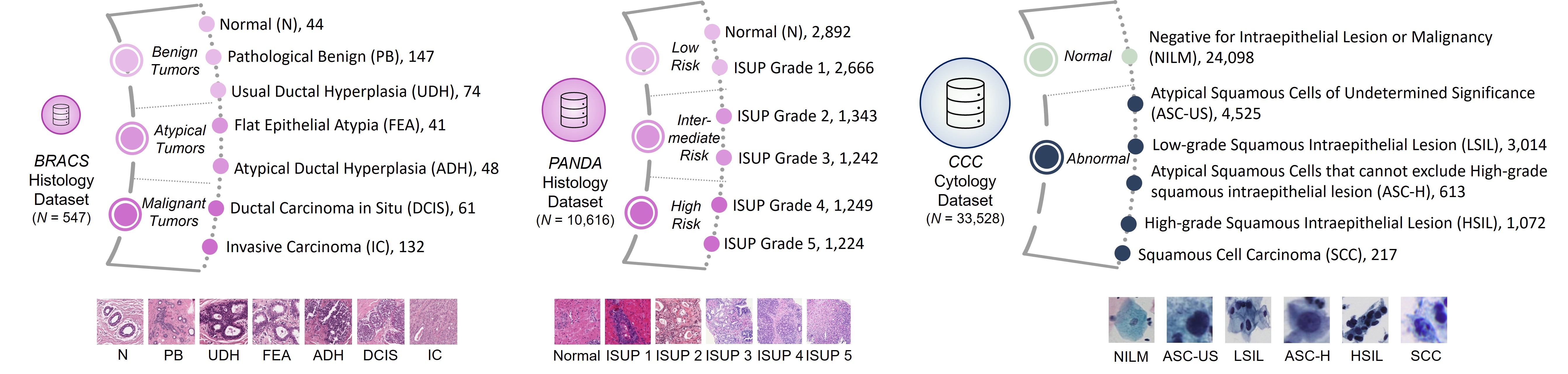
Our framework follows the hierarchy designed by the pathologist, as shown below:
Our HMIL framework adopts a dual-branch structure: a coarse branch for coarse-grained classification and a fine branch for fine-grained classification. Between this dual-branch structure, we introduce hierarchical alignment at both instance and bag levels to better guide the learning process. At the instance level, both branches utilize class-wise attention-based MIL to introduce the foundation of hierarchical information, and the hierarchical instance matching module aligns the fine branch’s class-wise attention with the coarse branch’s class-wise attention through a fine-to-coarse similarity constrain. At the bag level, the hierarchical bag alignment module ensures fine-to-coarse prediction consistency by aligning the predictions of both branches. Moreover, we incorporate supervised contrastive learning to strengthen the discriminative capability of the fine branch by maximizing inter-class distances and minimizing intra-class variations. Recognizing that the broad knowledge provided by the coarse branch may not sufficiently guide fine-grained classification, we introduce a dynamic weighting strategy to balance the influence between the coarse and fine branches during training.
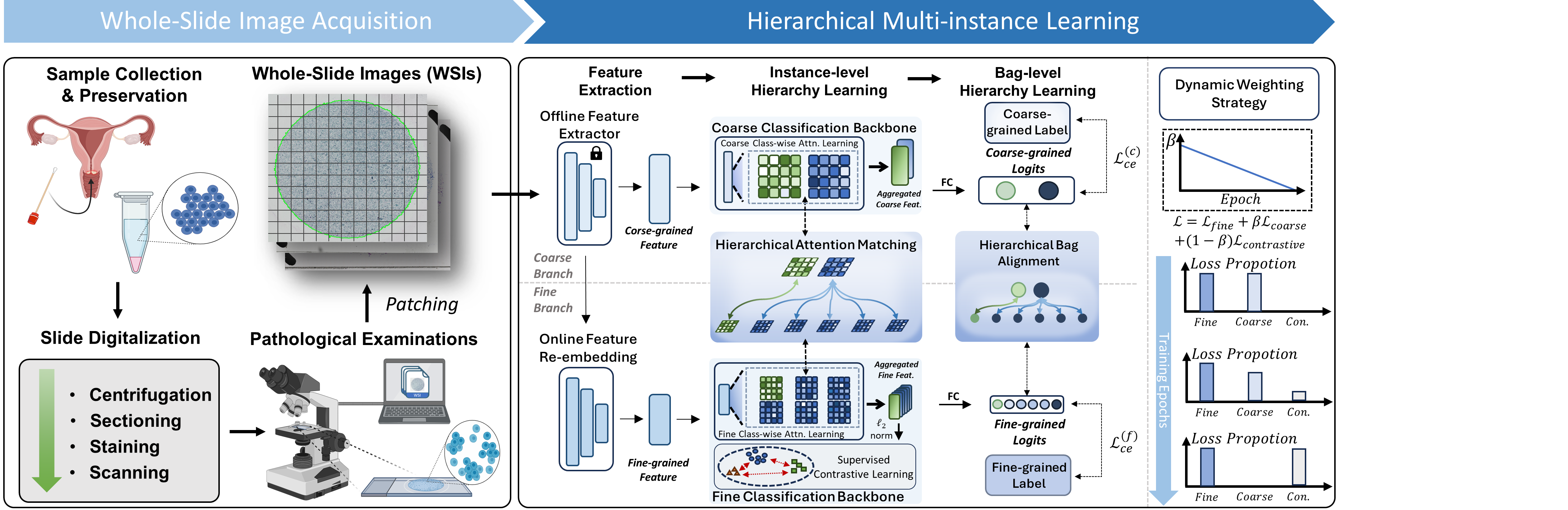
For the technical detail, please refer to the original paper.
Downloads
Full Paper: click here
PyTorch Code: Click here
Reference
@ARTICLE{jin2024hmil,
author={Jin, Cheng and Luo, Luyang and Lin, Huangjing and Hou, Jun and Chen, Hao},
journal={IEEE Transactions on Medical Imaging},
title={HMIL: Hierarchical Multi-Instance Learning for Fine-Grained Whole Slide Image Classification},
year={2025},
volume={44},
number={4},
pages={1796-1808},
doi={10.1109/TMI.2024.3520602}}