Abstract
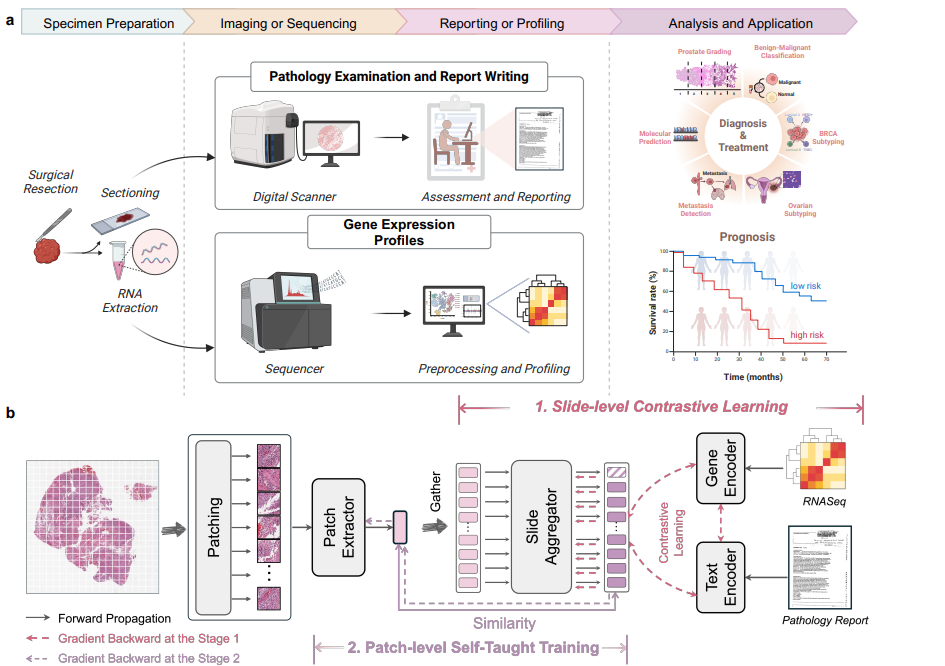
Remarkable strides in computational pathology have been made in the task-agnostic foundation model that advances the performance of a wide array of downstream clinical tasks. Despite the promising performance, there are still several challenges. First, prior works have resorted to either vision-only or vision-captions data, disregarding invaluable pathology reports and gene expression profiles which respectively offer distinct knowledge for versatile clinical applications. Second, the current progress in pathology FMs predominantly concentrates on the patch level, where the restricted context of patch-level pretraining fails to capture whole-slide patterns. Here we curated the largest multimodal dataset consisting of H\&E diagnostic whole slide images and their associated pathology reports and RNA-Seq data, resulting in 26,169 slide-level modality pairs from 10,275 patients across 32 cancer types. To leverage these data for CPath, we propose a novel whole-slide pretraining paradigm which injects multimodal knowledge at the whole-slide context into the pathology FM, called Multimodal Self-TAught PRetraining (mSTAR). The proposed paradigm revolutionizes the workflow of pretraining for CPath, which enables the pathology FM to acquire the whole-slide context. To our knowledge, this is the first attempt to incorporate multimodal knowledge at the slide level for enhancing pathology FMs, expanding the modelling context from unimodal to multimodal knowledge and from patch-level to slide-level. To systematically evaluate the capabilities of mSTAR, extensive experiments including slide-level unimodal and multimodal applications, are conducted across 7 diverse types of tasks on 43 subtasks, resulting in the largest spectrum of downstream tasks. The average performance in various slide-level applications consistently demonstrates significant performance enhancements for mSTAR compared to SOTA FMs.
Schematic Diagram of the Proposed Method
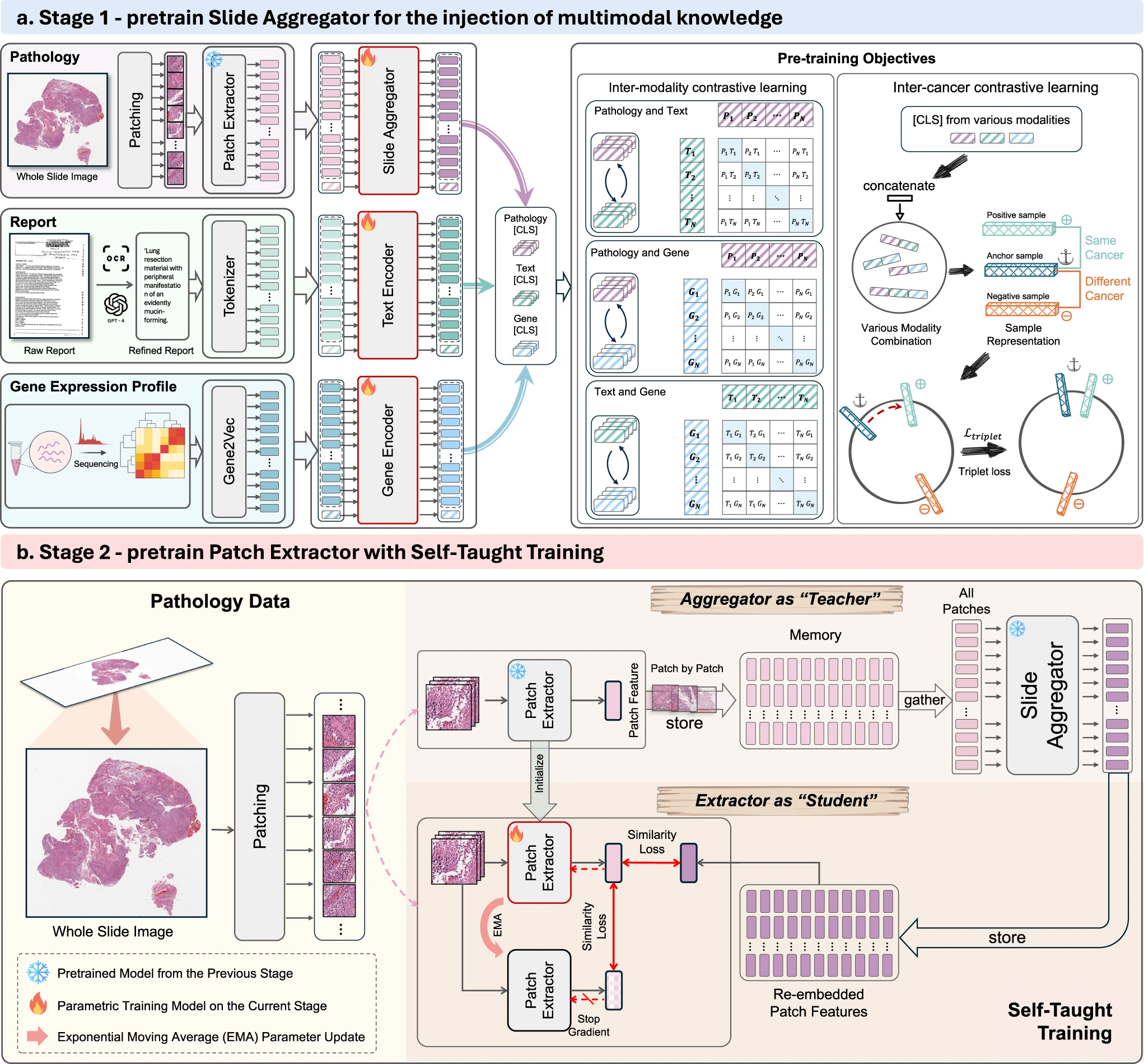
To utilize multimodal knowledge at the whole-slide context for enhancing the pathology foundation model, we propose a whole-slide pretraining paradigm consisting of two-stage pretraining, as shown in the following figure. In the first stage, we aim to inject multimodal knowledge into the slide aggregator by contrastive learning, including inter-modality contrastive learning (following CLIP) and inter-cancer contrastive learning. In the second stage, to seamlessly propagate multimodal knowledge at the slide-level context into the patch extractor, we leverage the slide aggregator pretrained in the first stage, serving as a “Teacher” model, to supervise the pretraining of the patch extractor, termed Self-Taught training. In this way, multimodal knowledge of the whole-slide context can be injected into the pathology FM.
For the technical details, please refer to the original paper.
Downloads
Full Paper: click here
Supplementary: click here
Peer Review History: click here
PyTorch Code: click here
Reference
@article{xu2025multimodal,
title={A multimodal knowledge-enhanced whole-slide pathology foundation model},
author={Xu, Yingxue and Wang, Yihui and Zhou, Fengtao and Ma, Jiabo and Jin, Cheng and Yang, Shu and Li, Jinbang and Zhang, Zhengyu and Zhao, Chenglong and Zhou, Huajun and others},
journal={Nature Communications},
volume={16},
pages={11406},
year={2025}
}