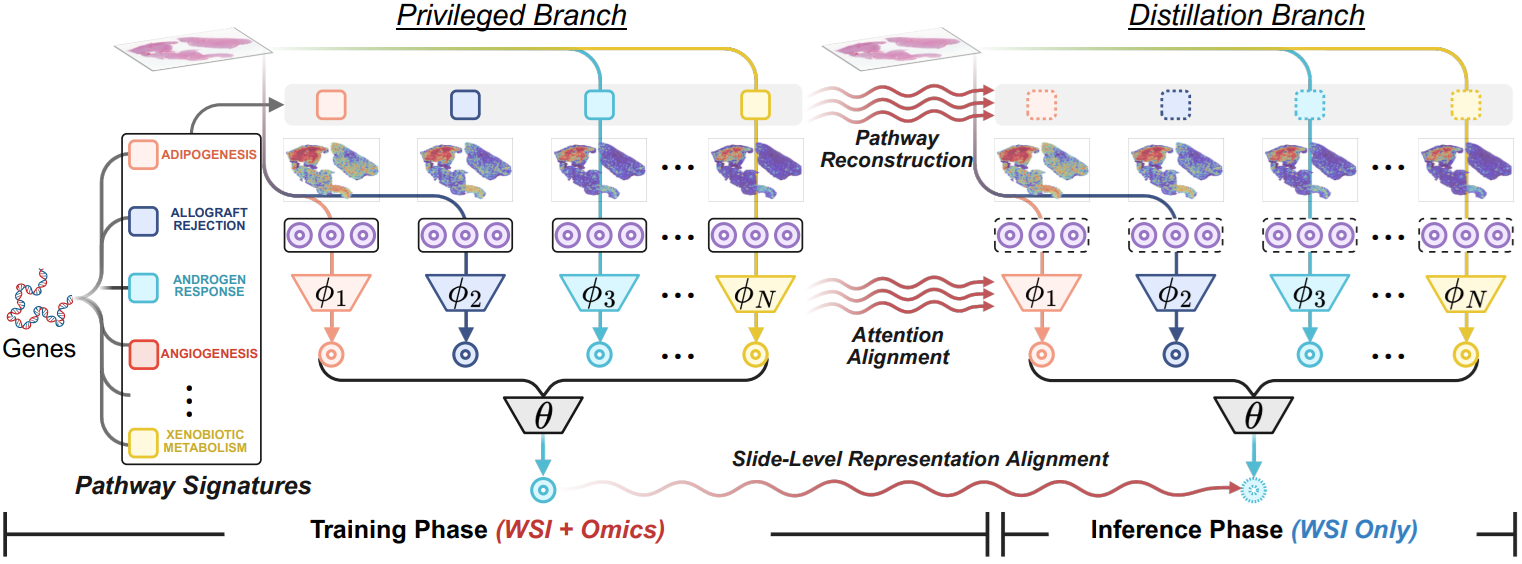
Abstract
Precision oncology requires accurate molecular insights, yet obtaining these directly from genomics is costly and time-consuming for broad clinical use. Predicting complex molecular features and patient prognosis directly from routine whole-slide images (WSI) remains a major challenge for current deep learning methods. Here we introduce PathLUPI, which uses transcriptomic privileged information during training to extract genome-anchored histological embeddings, enabling effective molecular prediction using only WSIs at inference. Through extensive evaluation across 49 molecular oncology tasks using 11,257 cases among 20 cohorts, PathLUPI demonstrated superior performance compared to conventional methods trained solely on WSIs. Crucially, it achieves AUC ≥ 0.80 in 14 of the biomarker prediction and molecular subtyping tasks and C-index ≥ 0.70 in survival cohorts of 5 major cancer types. Moreover, PathLUPI embeddings reveal distinct cellular morphological signatures associated with specific genotypes and related biological pathways within WSIs. By effectively encoding molecular context to refine WSI representations, PathLUPI overcomes a key limitation of existing models and offers a novel strategy to bridge molecular insights with routine pathology workflows for wider clinical application.
For the technical detail, please refer to the original paper.
Downloads
Arxiv Preprint: click here
PyTorch Code: click here
Reference
@article{jin2025pathlupi,
title={Genome-Anchored Foundation Model Embeddings Improve Molecular Prediction from Histology Images},
author={Jin, Cheng and Zhou, Fengtao and Yu, Yunfang and Ma, Jiabo and Wang, Yihui and Xu, Yingxue and Zhou, Huajun and Jiang, Hao and Luo, Luyang and Mao, Luhui and He, Zifan and Zhang, Xiuming and Zhang, Jing and Chan, Ronald and Yao, Herui and Chen, Hao},
journal={arXiv preprint arXiv:2506.19681},
year={2025}
}